Pilot projects are ideal for species new to GBS or for species with industry-standard protocols that want to move away from a frequent-cutter enzyme to improve sequencing costs. The pilot project enables a researcher to validate a subset of their sample collection with one double-digest to assess the number of informative markers and to estimate genetic diversity across recovered loci before committing to a full GBS project. The deep sequencing in a pilot project allows for scaling the amount of sequencing of your full sample set based on the desired number of SNPs and the overall project budget.
Because there is not a way to predict the SNP outcome for a given depth of sequencing, nearly all of our GBS clients with new species elect to begin with a pilot project. Pilots are bypassed if a client has a previous GBS data-set with a specific restriction enzyme they have been using for their specie.
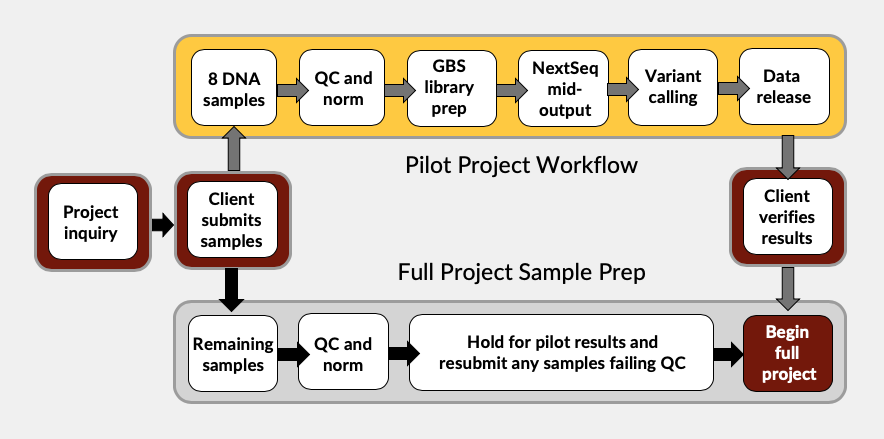
Pilot Workflow
Client selects 8 samples that represent the diversity of the population(s), or parents and the offspring of the crosses, to test with one double-digest and sequence deeply to 4 million reads/sample. Restriction enzymes selected are based on in silico digests, or if no reference genome is available, genome size plus the desired number of markers for the study. As with most NGS applications: more markers = more sequencing = greater costs.
Client selects 8 samples that represent the diversity of the population(s), or parents and the offspring of the crosses, to test with one double-digest and sequence deeply to 4 million reads/sample. Restriction enzymes selected are based on in silico digests, or if no reference genome is available, genome size plus the desired number of markers for the study. As with most NGS applications: more markers = more sequencing = greater costs.
Pilot analysis
Our bioinformatics team will analyze the pilot data three times:
- Using the full sequence data at 4 million reads/sample
- Sub-sampled down to 2 million reads/sample
- At 1 million reads/sample
Pilot deliverables
A full set of deliverables (raw fastQCs, vcf files, and summary report) are provided for each analysis to enable the client to determine which read depth meets their project goals and budget. Once the client verifies the sequencing depth, library prep will begin on the full sample set using the protocol determined in the pilot project.
Pilot timeline
It will take approximately 8-10 weeks for the pilot project’s vcf files and summary report to be released. Running parallel to the pilot project, clients can elect to submit samples for their full GBS project to begin QC and sample normalization.
Start a Pilot
Share with us your species details and genotyping needs using our GBS project inquiry form, and our GBS team will contact you to discuss experimental design.
Inquire for pricing. 8 samples are sequenced deeply to 4 - 12 million reads/sample (90 million reads total for the pilot project) and analyzed at 1, 2, and 4 million reads/sample or greater when possible. Client determines the sequencing depth for their full GBS project. Contact gbs@umn.edu.
Submission
Samples can be dropped off at one of our campus locations.
- 1-210 Cancer & Cardiovascular Research Building (Minneapolis campus)
- 20 Snyder Hall (St. Paul campus)
If shipping samples, the following address should be used.
Please send the tracking information to gbs@umn.edu.
UMN Genomics Center
ATTN: Corbin Dirkx
3510 Hopkins Place N.
Building 4 Suite W402
Oakdale, MN 55128
612-625-7736
Deliverables
Data Release
There are four options for transferring data from the UMGC to clients: 1) delivery to the Minnesota Supercomputing Institute’s (MSI) high-performance file system, 2) download from a secure website, 3) download with Globus, or 4) shipment on an external hard drive. Please indicate your data delivery preference when placing an order for sequencing.
1. MSI storage
Internal clients have their data released to MSI's Shared User Resource Facility Storage (SURFS). Delivered data will be located in the "data_delivery" folder in your group's folder on MSI's primary filesystem (home/GROUP/data_delivery/umgc). MSI does not charge for SURFS storage costs, but files expire and are removed one year after they've been delivered. Files should be copied to other MSI storage locations such as Tier2, Tier3, or your group's "shared" folder before they expire.
2. Web download
Internal clients that opt-out of MSI storage and external clients can download their data from a secure website using either a web browser or a command-line download tool, complete instructions are provided in an email from the UMGC. The client’s data is available for download for 30 days, after which the data will be removed from the data download website and the client takes responsibility for storing the data.
3. Globus
Internal and External clients can use Globus to download their data. This is the recommended method for external clients to download large datasets.
4. Hard drive
External clients may have data shipped on a hard drive purchased by the UMGC and invoiced to the client at a cost of $250 per hard drive.
Data Recovery
The UMGC archives most customer data for a year and some datasets are retained for 5 years or more. If you need a dataset re-delivered email a request to next-gen@umn.edu to initiate data recovery. The UMGC does not provide any guarantee that data can be successfully recovered from the archive.