RNA Sequencing (RNA-Seq) has become the gold standard method for genome-wide gene expression analysis in different biological conditions, as well as the detection of splice variants, transcriptome analysis, and the discovery non-coding RNAs. We provide several different library kit options for RNA-Seq library creation to accommodate varying sample types, including kits for low input and partially degraded samples, as well as options for polyA enrichment or ribosomal reduction. The UMGC offers a complete RNA-Seq workflow starting from RNA extraction through RNA QC analysis, and in partnership with MSI, analysis of RNA-Seq data through streamlined pipelines.
Please see our PacBio service to sequence full-length transcripts up to 10kb or longer and our Single-cell service for gene expression profiling of single cells.
Charter Service
Charter Service is our low-cost RNA-Seq service at $171.84/sample (UMN rate, 20 M reads) that provides a monthly shared sequencing run that departs. RNA samples are submitted in increments of 8 with a 16-sample minimum and must pass QC by the 1st of the previous month to start either the TruSeq Stranded mRNA kit or the Pico Mammalian v2 kit. Samples passing QC at the scheduled validation deadline are guaranteed to have data released in 4 weeks.
Batching RNA-Seq projects into a shared, scheduled monthly run enables us to offer a competitive RNA-Seq rate with a guaranteed 1-month turnaround time. Samples are sequenced on a 150 PE flow cell.
Please see the Charter Service calendar for upcoming submission deadlines and data release turnaround times.
Standard Service
Unlike the Charter Service, our Standard RNA-Seq Service has no restrictions regarding minimum number of samples, depth or read length, QC cut-off dates, and includes any of the library prep kits available at the UMGC. However, clients who elect for this service will follow our standard sequencing queue turnaround time and will see slightly longer turnaround times than the Charter Service. This is an a-la-carte service where the cost of the library prep and sequencing are priced separately, where clients purchase sequencing in lane increments.
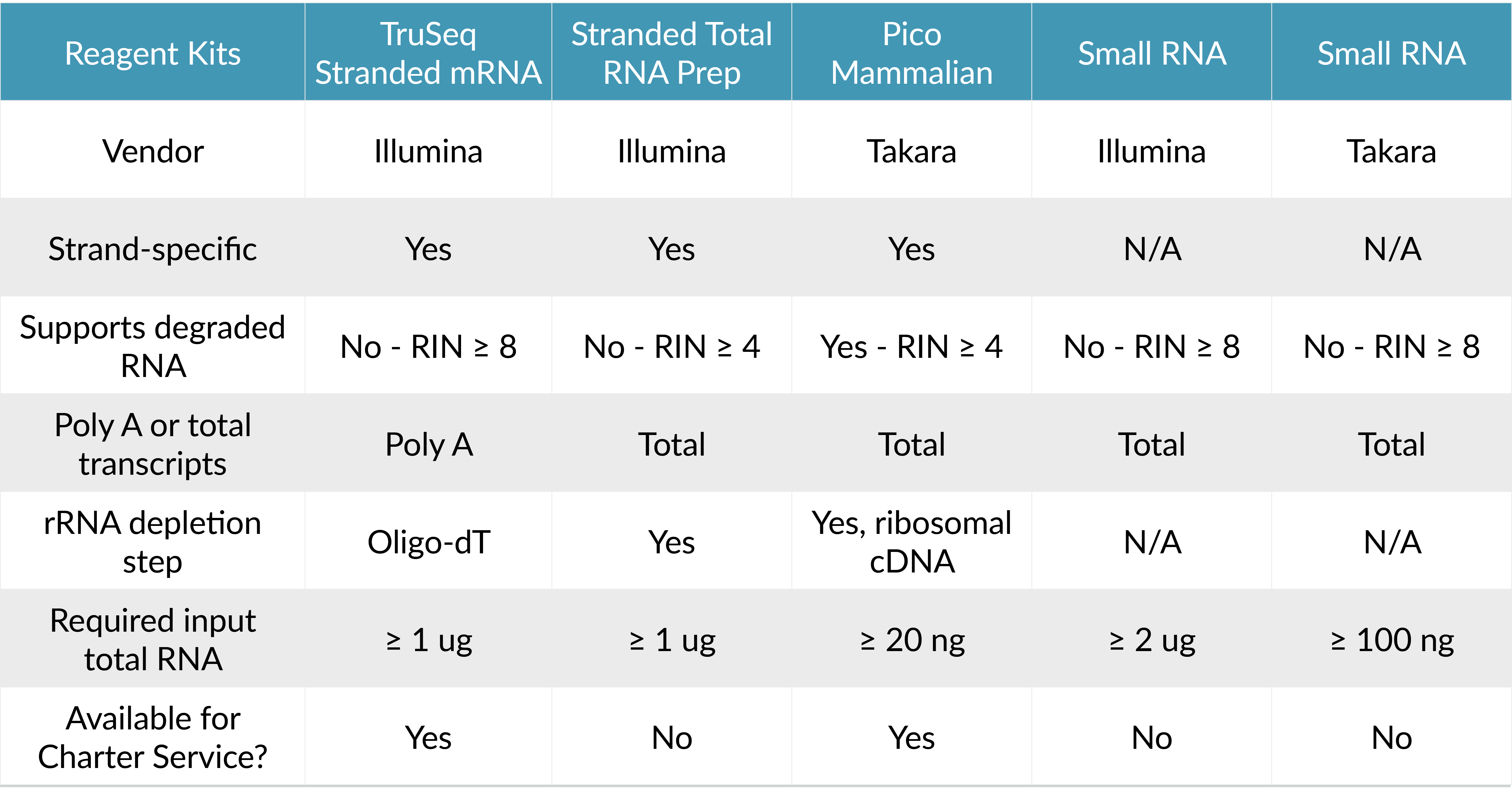
Comparison of RNA-Seq kits available at the UMGC. 1) Illumina Stranded Total RNA Prep includes the Illumina Ribo-Zero Plus rRNA Depletion Kit, which provides efficient removal of ribosomal RNA from multiple species, including human, mouse, rat, and bacteria. 2) UMGC sample submission requirements.
Questions?
For questions or a quote, please contact Elyse Froehling at next-gen@umn.edu.
UMN Rates
| Charter Service1 | Savings Tier | Speed Tier | Priority Tier |
|---|---|---|---|
| 20 million reads. 150 PE sequencing. | $171.84 | N/A | N/A |
| 40 million reads. 150 PE sequencing. | $222.39 | N/A | N/A |
| Illumina kit options2 | Savings Tier | Speed Tier | Priority Tier |
|---|---|---|---|
| mRNA. TruSeq Stranded. | $121.22 | $211.83 | $274.22 |
| Turnaround time. | 13 - 18 days | 7 - 13 days | 5 - 7 days |
| Total RNA. Stranded. Lig w RZ Plus. | $153.14 | $241.88 | $345.37 |
| Turnaround time. | 13 - 18 days | 7 - 13 days | 5 - 7 days |
| Takara kit options2 | Savings Tier | Speed Tier | Priority Tier |
|---|---|---|---|
| SMARTer Pico Mammalian. V2. | $110.30 | $197.30 | $227.57 |
| Turnaround time. | 13 - 18 days | 7 - 13 days | 5 - 7 days |
| Small RNA. | $157.20 | $257.16 | $323.91 |
| Turnaround time. | 13 - 18 days | 7 - 13 days | 5 - 7 days |
Notes
1. Charter Service: Charter Service is a shared, scheduled sequencing run on the 1st of the month with submission timeline requirements. Samples are submitted in batches of 8 with a 16-sample minimum. Posted rate includes library prep + sequencing. Kit options are the Illumina TruSeq Stranded mRNA or the Takara SMARTer Pico Mammalian. V2. RNA sample QC (Ribogreen + RNA sizing) is additional.
2. Library Creation: Standalone library creation rate. Please see our sequencing rates for your own dedicated lane and read length and depth. Standard sequencing queue turnaround applies. RNA sample QC (Ribogreen + RNA sizing) is additional.
Tiers: Turnaround time (TAT) is in business days. TAT is for library prep only and does not factor in the TAT for sequencing or other companion services. Availability for the Priority Tier is limited; contact next-gen@umn.edu to see if this is a current option. More information about Pricing Tiers.
External Rates
| Charter Service1 | Savings Tier | Speed Tier | Priority Tier |
|---|---|---|---|
| 20 million reads. 150 PE sequencing. | $210.30 | N/A | N/A |
| 40 million reads. 150 PE sequencing. | $270.56 | N/A | N/A |
| Illumina kit options2 | Savings Tier | Speed Tier | Priority Tier |
|---|---|---|---|
| mRNA. TruSeq Stranded. | $149.89 | $250.61 | $319.83 |
| Turnaround time. | 13 - 18 days | 7 - 13 days | 5 - 7 days |
| Total RNA. Stranded. Lig w RZ Plus. | $189.38 | $288.00 | $402.99 |
| Turnaround time. | 13 - 18 days | 7 - 13 days | 5 - 7 days |
| Takara kit options2 | Savings Tier | Speed Tier | Priority Tier |
|---|---|---|---|
| SMARTer Pico Mammalian. V2. | $136.40 | $233.09 | $266.71 |
| Turnaround time. | 13 - 18 days | 7 - 13 days | 5 - 7 days |
| Small RNA. | $194.40 | $305.52 | $379.61 |
| Turnaround time. | 13 - 18 days | 7 - 13 days | 5 - 7 days |
Notes
1. Charter Service: Charter Service is a shared, scheduled sequencing run on the 1st of the month with submission timeline requirements. Samples are submitted in batches of 8 with a 16-sample minimum. Posted rate includes library prep + sequencing. Kit options are the Illumina TruSeq Stranded mRNA or the Takara SMARTer Pico Mammalian. V2. RNA sample QC (Ribogreen + RNA sizing) is additional.
2. Library Creation: Standalone library creation rate. Please see our sequencing rates for your own dedicated lane and read length and depth. Standard sequencing queue turnaround applies. RNA sample QC (Ribogreen + RNA sizing) is additional.
Tiers: Turnaround time (TAT) is in business days. TAT is for library prep only and does not factor in the TAT for sequencing or other companion services. Availability for the Priority Tier is limited; contact next-gen@umn.edu to see if this is a current option. More information about Pricing Tiers.
Submission
How to Order
1. Please contact next-gen@umn.edu for project specifications.
2. Once project details are finalized, complete the appropriate submission form for submitting samples or submitting client-made libraries and email to next-gen@umn.edu.
Illumina Sequencing Request Form (Libraries)
Illumina Sequencing Request Form (Samples)
Shipping Instructions
Samples should be frozen and shipped on dry ice in a 96-well plate. We recommend using plate tape to seal the wells. Place the plate inside of a plastic bag prior to placing on dry ice. Please give advance notice of submission date and time so staff can be prepared to receive samples. If shipping samples from outside the University of Minnesota, ship via express shipping carrier on dry ice to the address below.
Samples can be dropped off at one of our campus locations
- 1-210 Cancer & Cardiovascular Research Building (Minneapolis campus)
- 20 Snyder Hall (St. Paul campus)
Samples can be shipped to the following address:
Please send the tracking information to next-gen@umn.edu.
UMN Genomics Center
ATTN: NGS Staff
3510 Hopkins Place N.
Building 4 Suite W402
Oakdale, MN 55128
612-625-7736
Deliverables
Data Release
There are four options for transferring data from the UMGC to clients: 1) delivery to the Minnesota Supercomputing Institute’s (MSI) high-performance file system, 2) download from a secure website, 3) download with Globus, or 4) shipment on an external hard drive. Please indicate your data delivery preference when placing an order for sequencing.
1. MSI storage
Internal clients have their data released to MSI's Shared User Resource Facility Storage (SURFS). Delivered data will be located in the "data_delivery" folder in your group's folder on MSI's primary filesystem (home/GROUP/data_delivery/umgc). MSI does not charge for SURFS storage costs, but files expire and are removed one year after they've been delivered. Files should be copied to other MSI storage locations such as Tier2, Tier3, or your group's "shared" folder before they expire.
2. Web download
Internal clients that opt-out of MSI storage and external clients can download their data from a secure website using either a web browser or a command-line download tool, complete instructions are provided in an email from the UMGC. The client’s data is available for download for 30 days, after which the data will be removed from the data download website and the client takes responsibility for storing the data.
3. Globus
Internal and External clients can use Globus to download their data. This is the recommended method for external clients to download large datasets.
4. Hard drive
External clients may have data shipped on a hard drive purchased by the UMGC and invoiced to the client at a cost of $250 per hard drive.
Data Recovery
The UMGC archives most customer data for a year and some datasets are retained for 5 years or more. If you need a dataset re-delivered email a request to next-gen@umn.edu to initiate data recovery. The UMGC does not provide any guarantee that data can be successfully recovered from the archive.
FAQ - Charter Service
How do I reserve my seats and how many seats can I request?
Reserve your seats by emailing your request to next-gen@umn.edu. You can request as many seats as you need as long as you are purchasing a minimum of 2 seats (16 samples) and groups of 8 samples thereafter. The flight has a maximum of 48 seats (384 samples). Refer to Sequencer Home to see the availability of the next run.
What samples are acceptable?
We accept eukaryotic total RNA samples only for this service. No bacterial or Ribozero service is currently being offered.
Do you perform QC on samples?
Yes, the UMGC provides QC services for all RNA-seq samples. Samples MUST be pre-validated before they are accepted for the Charter Service.
What is the deadline for submission?
2 weeks (10 working days) prior to the flight date. This allows UMGC the opportunity to perform sample QC and obtain replacement samples, if necessary. Refer to Sequencer Home to see the availability of the next run.
Can I request Express Service for Charter-Seq?
There is no Express service. The dates are set.
What if I miss the deadline for submission?
You will have the option of having your samples processed using our standard RNA-seq service and purchasing a separate lane of sequencing. Or, you can wait until the next Charter Service flight.
I can't seem to get on a Charter-Seq run because they fill up too fast.
We have the ability to increase the frequency of Charter Service runs, and will do so if we find that demand justifies it.
FAQ - Standard Service
What is the turnaround time?
Generally, turnaround is 6-8 weeks for standard NGS projects. However, please contact next-gen@umn.edu for specific project turnaround time.