NanoString GeoMx Digital Spatial Profiler
NanoString’s GeoMx™ Digital Spatial Profiler (DSP) combines standard immunofluorescence techniques with digital optical barcoding technology to perform highly multiplexed, spatially resolved profiling experiments. In a single reaction, the DSP technology performs whole slide imaging with up to four fluorescent stains to capture tissue morphology and select regions of interest for high plex profiling. The DSP chemistry enables spatially resolved high plex profiling of RNA and protein targets on just two serial sample sections, with no-destructive tissue processing.
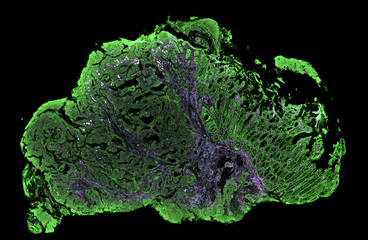
- Minimum sample: protein or RNA analysis from formalin-fixed paraffin-embedded (FFPE) or fresh-frozen section.
- Morphological context: whole-slide 4 color imaging to guide profiling.
- Digital quantitation: up to 6 logs (base 10) dynamic range.
- Preserve precious samples with non-destructive processing.
Key applications include immunology, immuno-oncology, neurodegeneration, and neuroinflammation. Many different applications can be supported on GeoMx Digital Spatial Profiler and the content menu is continuously expanding. See the NanoString website to explore available RNA and protein assays, including the recently released GeoMx Whole Transcriptome Atlas.
Workflow
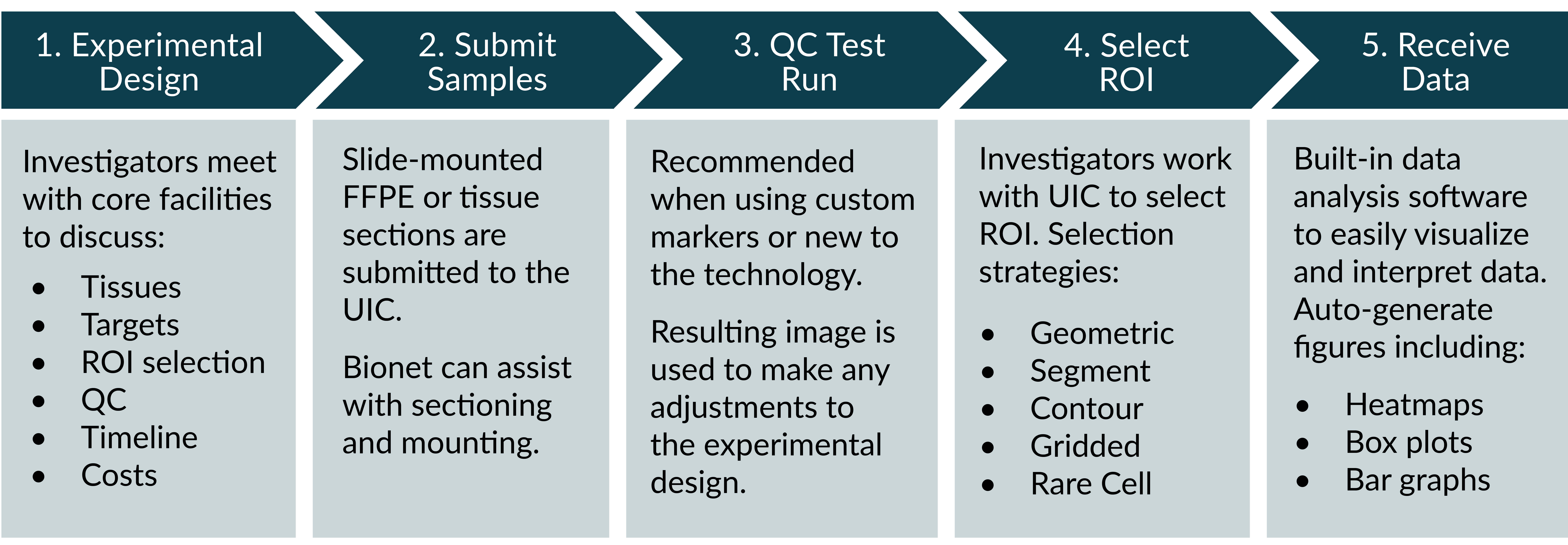
The NanoString GeoMx Service is supported by the collaborative efforts of three core facilities at the University of Minnesota: Bionet, UIC, and the UMGC.
- UMN Biological Material Procurement Network (Bionet) provides optional assistance with tissue sectioning and mounting of FFPE or fresh frozen tissue. BioNet has multiple locations on-campus.
- University Imaging Center (UIC) stains tissue slides with pre-mixed biological probes and fluorescently-labeled markers, works with investigators on ROI selection, and uses UV exposure to release DSP barcode tags from ROI.
- UMN Genomics Center (UMGC) quantifies released tags on the nCounter or on an Illumina next-generation sequencer and counts are mapped back to tissue location to yield a digital profile. The UMGC also provides consultation on the GeoMX DSP technology and assists with experimental design.
The UMGC and UIC are neighboring facilities located at 1-210 and 1-220 Cancer & Cardiovascular Research Building (CCRB).
Start a Project

Researchers will meet with Dr. Fernanda Rodriguez (UMGC), Grant Barthel (UIC), and Colleen Forster (Bionet) to discuss downstream research goals and align with the experimental design.
In this meeting, we will review available spatial platforms and if the client is ready, we will discuss the number and type of samples, targets of interest (RNA/protein), morphology markers, region of interest (ROI) selection strategies, QC, workflow, timelines, and general overall costs. A specific quote can be provided as per user request.
Because slides with samples for RNA projects need to be stained and processed within two weeks of being mounted, the spatialomics team and the researcher work closely together for seamless coordination. ROI selection is done by the user, via a Zoom meeting or in person, with Grant’s support.
At the data hand-off meeting, Dr. John Garbe (UMGC) and Grant will provide an overview of the GeoMx software with the user’s data. The user will receive an email with the link to the software and access information that can be shared with collaborators. More in-depth analysis is available with the MSI’s RIS group, please contact Dr. Christine Henzler.
Questions
As the first school in the Big Ten to receive the DSP instrument and adopt this novel technology, the UMGC and UIC scientists are able to use their technical experience to guide and inform researchers on the advantages and current limitations of the technology. Contact Dr. Fernanda Rodriquez at spatialomics@umn.edu with questions on the NanoString’s DSP platform or to schedule a consultation.
Join the UMN Spatialomics Group to connect with other UMN spatial researchers and experts in the spatialomics field.
UMN Rates
| Service | Savings Tier | Speed Tier | Priority Tier |
|---|---|---|---|
| GeoMx library prep processing. Per sample. | $16.53 | $31.42 | $61.21 |
| Turnaround time. | 3 to 6 days | 1 to 3 days | 1 to 2 days |
| GeoMx RNA. Multiples of 12 libraries. | $172.54 | N/A | N/A |
Notes
Tiers: Turnaround time (TAT) is in business days. The TAT is for the above-stated services and does not factor in the TAT for sequencing or other companion services. Availability for the Priority Tier is limited; contact spatialomics@umn.edu to see if this is a current option. More information about Pricing Tiers.
External Rates
| Service | Savings Tier | Speed Tier | Priority Tier |
|---|---|---|---|
| GeoMx library prep processing. Per sample. | $21.10 | $37.66 | $70.74 |
| Turnaround time. | 3 to 6 days | 1 to 3 days | 1 to 2 days |
| GeoMx RNA. Multiples of 12 libraries. | $220.10 | N/A | N/A |
Notes
Tiers: Turnaround time (TAT) is in business days. The TAT is for the above-stated services and does not factor in the TAT for sequencing or other companion services. Availability for the Priority Tier is limited; contact spatialomics@umn.edu to see if this is a current option. More information about Pricing Tiers.
Deliverables
Data Release
There are four options for transferring data from the UMGC to clients: 1) delivery to the Minnesota Supercomputing Institute’s (MSI) high-performance file system, 2) download from a secure website, 3) download with Globus, or 4) shipment on an external hard drive. Please indicate your data delivery preference when placing an order for sequencing.
1. MSI storage
Internal clients have their data released to MSI's Shared User Resource Facility Storage (SURFS). Delivered data will be located in the "data_delivery" folder in your group's folder on MSI's primary filesystem (home/GROUP/data_delivery/umgc). MSI does not charge for SURFS storage costs, but files expire and are removed one year after they've been delivered. Files should be copied to other MSI storage locations such as Tier2, Tier3, or your group's "shared" folder before they expire.
2. Web download
Internal clients that opt-out of MSI storage and external clients can download their data from a secure website using either a web browser or a command-line download tool, complete instructions are provided in an email from the UMGC. The client’s data is available for download for 30 days, after which the data will be removed from the data download website and the client takes responsibility for storing the data.
3. Globus
Internal and External clients can use Globus to download their data. This is the recommended method for external clients to download large datasets.
4. Hard drive
External clients may have data shipped on a hard drive purchased by the UMGC and invoiced to the client at a cost of $250 per hard drive.
Data Recovery
The UMGC archives most customer data for a year and some datasets are retained for 5 years or more. If you need a dataset re-delivered email a request to next-gen@umn.edu to initiate data recovery. The UMGC does not provide any guarantee that data can be successfully recovered from the archive.
