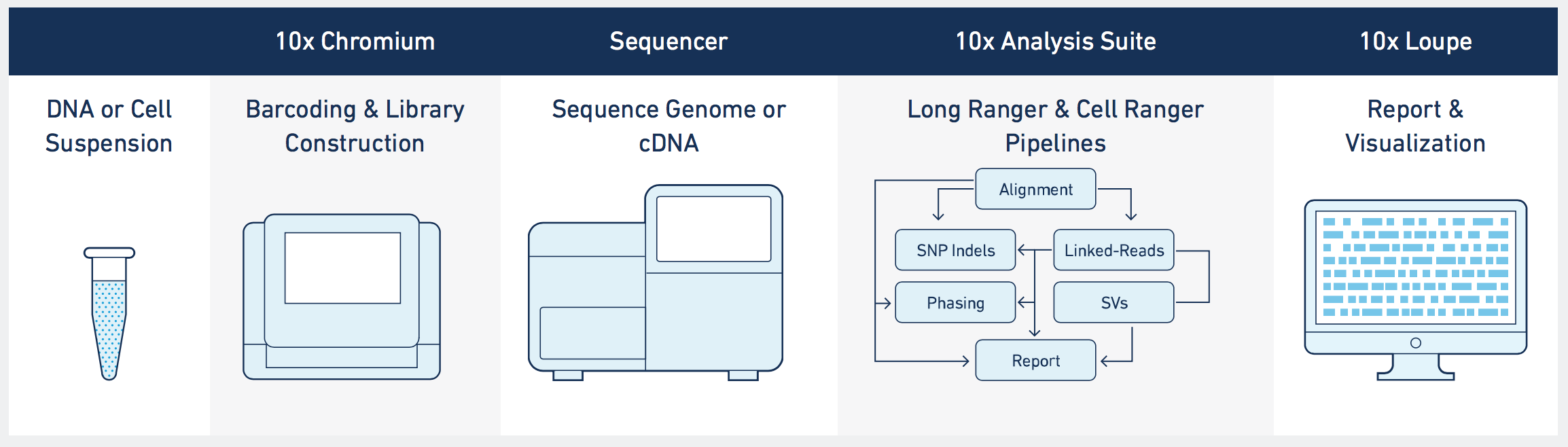
The UMN Genomics Center expands single-cell research capabilities with the addition of 10X Genomics Chromium X, a next-generation microfluidic platform for single-cell analysis. The granularity of the proprietary Next GEM technology allows researchers to interrogate individual cells and unmask rare cell types that may not be seen in bulk RNA sequencing methods.
The UMGC can handle a range of single-cell projects, from pilot experiments to the highest-throughput studies, and offers the full portfolio of validated 10X single-cell assays, to give researchers flexibility for any experimental strategy.
Key Features of Our Service
- Complete end-to-end workflow: cell suspension loading, 10X library creation, rigorous QC, sequencing, and analysis.
- Latest 10X instrument for analyzing hundreds to hundreds of thousands of cells in a single run.
- 10X libraries are sequenced on the production-scale NovaSeq X Plus for economical sequencing and project scalability.
- Support with experimental design and guidance on assay selection, including adding Feature Barcoding and Cellplex options, or our supported Cell Hashing and CITE-Seq protocols.
- Additional support services are available on-campus through the University Flow Cytometry Resource (UFCR) and the Minnesota Supercomputing Institute (MSI).
10X Single-cell Assays
The UMGC offers a wide and growing list of 10X single-cell assays to support a broad range of applications, including gene expression, chromatin accessibility, cell surface proteins, immune clonotype, antigen specificity, and CRISPR screens. We also support library preparation and sequencing of cDNA samples derived from 10X’s Visium spatial solution.
Workflow
Consult: We request a project planning meeting to review experimental goals and provide guidance on experimental design. See the above Guidelines tab for more info. We can also arrange for a dry run to optimize your cell preparation for the workflow before going into full project with sequencing.
Cell Prep: The 10X Genomics Cell Preparation Guide describes best practices and methods, but researchers should optimize their preparation protocol for their particular sample type. For cell samples needing sorting, the UMGC offers a MACSQuant Tyto cell sorter as a DIY instrument.
Two new commercially available kits improve the sample preparation workflow: 1) the Fixed RNA Profiling kit uses formaldehyde fixation to preserve samples at the point of collection, enabling sample batching for flexibility, and 2) the Nuclei Isolation kit enables analysis of frozen archival tissue.
Capture and Library Prep: We house the latest 10X Chromium X instrument. Up to eight samples can be processed per run allowing for the capture of up to 80,000 cells per run (cell capture efficiency ~65%). The UMGC offers all the 10X single-cell assays to give researchers the widest variety of experiments.
Sequencing: The UMGC uses a MiSeq QC run to check the number of targeted single cells before going into full-scale sequencing on the NovaSeq X Plus. The total amount of sequencing needed for a project is determined by the type of single-cell assay and the number of cells captured.
Data Delivery: Our 10X Genomics single-cell services include analysis on Gene Expression datasets using the Cellranger Count pipeline. The provided Cellranger “outs” folder will include an HTML report, a .cloupe file for use with the interactive Loupe Browser, and data files containing gene expression count data, as well as the FASTQ sequence files.
For analysis and interpretation of the Cellranger Count output, we recommend contacting the Research Informatics Solutions group at the Minnesota Supercomputing Institute for support.
Start a Project
Whether a researcher is new to single-cell sequencing or one of our experienced users, we can help determine which 10X single-cell assay will best meet your experimental goals. The UMGC requests that researchers contact us at single-cell@umn.edu for consultation on all incoming single-cell projects.
UMN Rates
| Capture | Savings Tier | Speed Tier | Priority Tier |
|---|---|---|---|
| 10X capture and reagents. No chip. | $2,753.62 | $3,051.83 | $4,110.93 |
| Turnaround time. | 5 - 9 days | 4 - 7 days | 2 - 4 days |
| 10X capture. No chip. | $2,615.12 | $2,807.19 | $3,677.79 |
| Turnaround time. | 0 - 1 days | 0 - 1 days | 0 - 1 days |
| 10X Multiome capture and reagents. No chip. | $4,218.11 | $4,824.11 | $6,253.53 |
| Turnaround time. | 6 - 10 days | 3 - 7 days | 3 - 4 days |
| Library Prep | Savings Tier | Speed Tier | Priority Tier |
|---|---|---|---|
| 10X SC GEX library with QC. Sample 1. | $863.49 | $962.71 | $1,205.67 |
| Turnaround time. | 10 - 16 days | 6 - 10 days | 4 - 6 days |
| 10X SC GEX library with QC. Samples 2-8. | $263.90 | $314.26 | $410.66 |
| Turnaround time. | 10 - 16 days | 6 - 10 days | 4 - 6 days |
| 10X SC add-on library with QC. | $259.92 | $438.38 | $741.24 |
| Turnaround time. | 8 - 13 days | 5 - 8 days | 3 - 5 days |
| 10X SC ATAC library with QC. | $863.49 | $962.71 | $1,205.67 |
| Turnaround time. | 10 - 16 days | 6 - 10 days | 4 - 6 days |
| 10X SC Multiome library with QC. Sample 1. | $1,135.53 | $1,493.69 | $2,221.22 |
| Turnaround time. | 16 - 24 days | 10 - 16 days | 6 - 9 days |
| 10X SC Multiome library with QC. Samples 2-8. | $460.11 | $734.37 | $1,264.92 |
| Turnaround time. | 16 - 24 days | 10 - 16 days | 6 - 9 days |
| 10X SC VDJ library with QC. | $429.23 | $824.85 | $1,498.18 |
| Turnaround time. | 10 - 16 days | 6 - 10 days | 4 - 6 days |
| 10X SC cDNA and QC. | $138.50 | $244.64 | $433.14 |
| Turnaround time. | 5 - 8 days | 4 - 6 days | 2 - 3 days |
Notes
Tiers: Turnaround time (TAT) is in business days. TAT is for the above-stated services and does not factor in the TAT for sequencing or other companion services. Availability for the Priority Tier is limited; contact single-cell@umn.edu to see if this is a current option. More information about Pricing Tiers.
External Rates
| Capture | Savings Tier | Speed Tier | Priority Tier |
|---|---|---|---|
| 10X capture and reagents. No chip. | $3,619.91 | $3,813.84 | $4,914.44 |
| Turnaround time. | 5 - 9 days | 4 - 7 days | 2 - 4 days |
| 10X capture. No chip. | $3,446.41 | $3,524.61 | $4,415.78 |
| Turnaround time. | 0 - 1 days | 0 - 1 days | 0 - 1 days |
| 10X Multiome capture and reagents. No chip. | $5,391.96 | $5,891.02 | $7,475.99 |
| Turnaround time. | 6 - 10 days | 3 - 7 days | 3 - 4 days |
| Library Prep | Savings Tier | Speed Tier | Priority Tier |
|---|---|---|---|
| 10X SC GEX library with QC. Sample 1. | $1,067.77 | $1,178.29 | $1,447.93 |
| Turnaround time. | 10 - 16 days | 6 - 10 days | 4 - 6 days |
| 10X SC GEX library with QC. Samples 2-8. | $326.33 | $382.33 | $489.37 |
| Turnaround time. | 10 - 16 days | 6 - 10 days | 4 - 6 days |
| 10X SC add-on library with QC. | $321.41 | $519.66 | $856.23 |
| Turnaround time. | 8 - 13 days | 5 - 8 days | 3 - 5 days |
| 10X SC ATAC library with QC. | $1,067.77 | $1,178.29 | $1,447.93 |
| Turnaround time. | 10 - 16 days | 6 - 10 days | 4 - 6 days |
| 10X SC Multiome library with QC. Sample 1. | $1,404.17 | $1,802.50 | $2,610.49 |
| Turnaround time. | 16 - 24 days | 10 - 16 days | 6 - 9 days |
| 10X SC Multiome library with QC. Sample 2-8. | $568.96 | $873.87 | $1,463.23 |
| Turnaround time. | 16 - 24 days | 10 - 16 days | 6 - 9 days |
| 10X SC VDJ library with QC. | $530.78 | $970.31 | $1,718.22 |
| Turnaround time. | 10 - 16 days | 6 - 10 days | 4 - 6 days |
| 10X SC cDNA and QC. | $173.50 | $289.23 | $498.66 |
| Turnaround time. | 5 - 8 days | 4 - 6 days | 2 - 3 days |
Notes
Tiers: Turnaround time (TAT) is in business days. TAT is for the above-stated services and does not factor in the TAT for sequencing or other companion services. Availability for the Priority Tier is limited; contact single-cell@umn.edu to see if this is a current option. More information about Pricing Tiers.
Submission
A planning meeting is needed to start a single-cell project to review your experimental goals and provide guidance on experimental design, as well as go over project logistics like turnaround estimates, scheduling, suspension buffer, and cell concentration requirements (varies based on assay).
Please note: Researchers need to schedule with the UMGC the delivery of a dissociated suspension of live cells or nuclei:
- We must have advance notice to have someone on standby to receive your cell suspension and prepare the reagents for cell capture.
- Your samples should be processed as quickly as possible.
- Email single-cell@umn.edu a completed Sample Submission Form ahead of cell or nuclei suspension/s drop-off.
On the scheduled day of your capture, the UMGC staff requests to be kept apprised of your progress throughout your prep and notified if your delivery of cell or nuclei suspension/s on ice will be more than 30 minutes earlier or later than your appointment.
While we aim to be flexible and recognize that cell preparations can be labor and time-intensive, deliveries of cell suspensions more than 30 minutes outside of appointment times are subject to nonacceptance and cancellation of capture.
Contact: single-cell@umn.edu
Drop-off Location: 1-210 Cancer & Cardiovascular Research Building (CCRB, Minneapolis campus)
Resources
MSI Intro to Single Cell Genomics
This tutorial is a lecture that covers basic experimental design principles as they pertain to single cell genomics. Covers common technological platforms for collecting single cell data, basic experimental design, data quality control, and overviews of common software tools used for analyzing single cell data. The material will cover some of the common applications of single cell data and compare them to bulk data. The main applications will be for single cell RNAseq, but the principles would be appropriate to other types single cell genomics data
- Recording (March 12, 2024)
- Introduction to Single-cell Genomics Slides
- Next tutorial
Deliverables
Data Release
There are four options for transferring data from the UMGC to clients: 1) delivery to the Minnesota Supercomputing Institute’s (MSI) high-performance file system, 2) download from a secure website, 3) download with Globus, or 4) shipment on an external hard drive. Please indicate your data delivery preference when placing an order for sequencing.
1. MSI storage
Internal clients have their data released to MSI's Shared User Resource Facility Storage (SURFS). Delivered data will be located in the "data_delivery" folder in your group's folder on MSI's primary filesystem (home/GROUP/data_delivery/umgc). MSI does not charge for SURFS storage costs, but files expire and are removed one year after they've been delivered. Files should be copied to other MSI storage locations such as Tier2, Tier3, or your group's "shared" folder before they expire.
2. Web download
Internal clients that opt-out of MSI storage and external clients can download their data from a secure website using either a web browser or a command-line download tool, complete instructions are provided in an email from the UMGC. The client’s data is available for download for 30 days, after which the data will be removed from the data download website and the client takes responsibility for storing the data.
3. Globus
Internal and External clients can use Globus to download their data. This is the recommended method for external clients to download large datasets.
4. Hard drive
External clients may have data shipped on a hard drive purchased by the UMGC and invoiced to the client at a cost of $250 per hard drive.
Data Recovery
The UMGC archives most customer data for a year and some datasets are retained for 5 years or more. If you need a dataset re-delivered email a request to next-gen@umn.edu to initiate data recovery. The UMGC does not provide any guarantee that data can be successfully recovered from the archive.