PacBio Sequel II Sequencing

The UMN Genomics Center continues to expand its capabilities in third-generation, single-molecule sequencing by leveraging the PacBio Sequel II as a workhorse long read platform. We offer end-to-end PacBio services, starting with rigorous sample QC and library preparation all the way through data analysis which can even include microbial de novo genome assembly.
Based on well-established Single Molecule, Real-Time (SMRT) Sequencing technology, the Sequel II System generates HiFi reads which are both long (up to 30 kb) and highly accurate (>99.9%), enabling high-quality genome assemblies, structural variant detection, transcriptomics and isoform detection, and epigenetic information, while providing high consensus accuracy and uniform coverage.
The Sequel II represents a significant improvement in long-read sequencing. The current SMRT Cell 8M provides 8X more sequencing data output, as well as reduced project costs and timelines compared to the prior version of the system.
As a PacBio Certified Service Provider, the UMGC undergoes certification to demonstrate we generate high quality data and the longest reads possible across a range of SMRT sequencing applications:
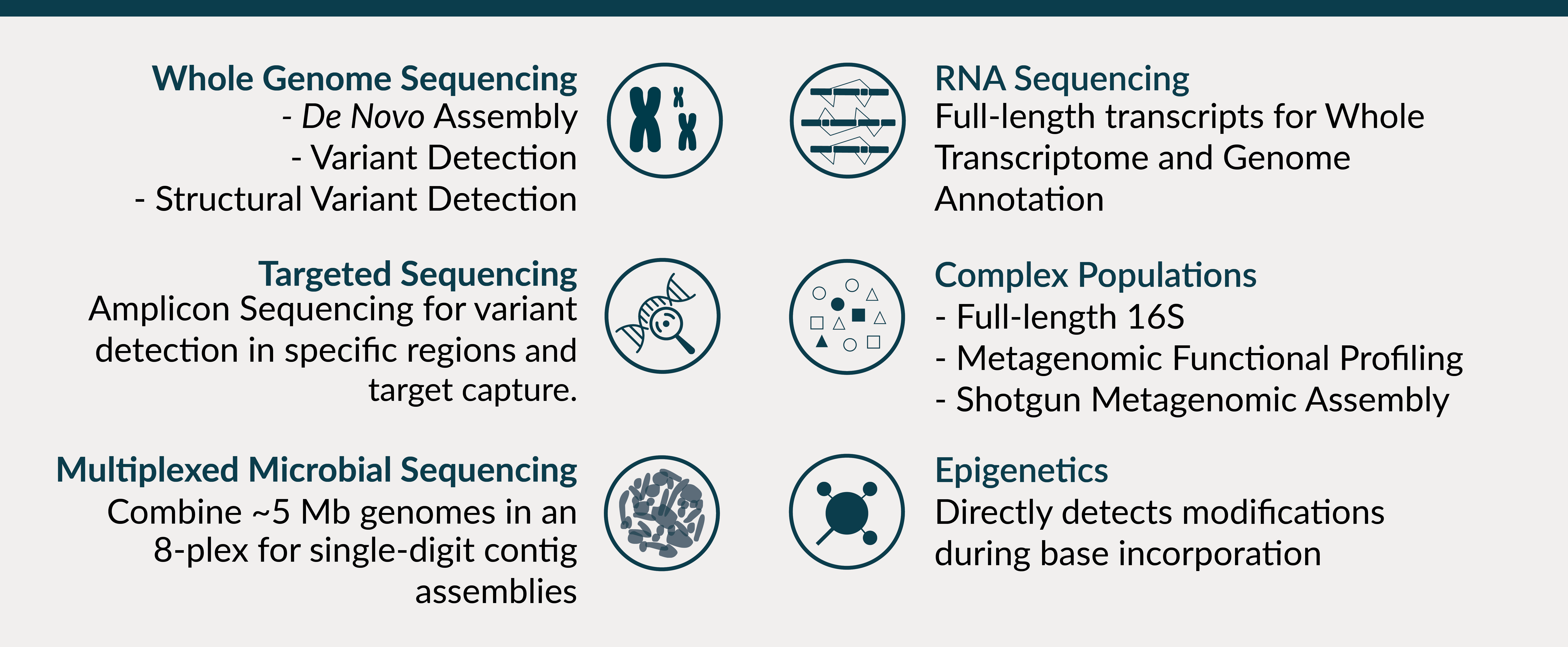
Output
Thanks to recent advances in sequencing chemistry and adaptive loading, the SMRT Cell 8M typically generates ~ 4-5M raw reads with flexible sequencing run times of up to 30 hours, yielding ~250-450 Gbp of raw data. As the sequencing polymerase makes successive passes along the same SMRT bell template, random errors present in each subread can be corrected to achieve high yields of >QV20 (99%) without sacrificing read length.

- Up to 160 Gb when using the Continuous Long Read (CLR) mode for de novo assembly and structural variant detection. By size selecting library fragments ≥30 kb, exceptionally long reads can be achieved with some > 150 kb.
- Up to ~500 Gb when using Circular Consensus Sequencing (CSS, HiFi) mode with 10 - 15 Gb after CSS and filtering for highly accurate amplicon and whole transcriptome sequencing projects.
Benefits
- Long Reads: With HiFi read lengths up to 25 kb, investigators can readily assemble complete genomes and sequence full-length transcripts. Long read lengths can resolve repetitive regions that are difficult for short-read technologies.
- High Accuracy: >99.999% consensus accuracy is achieved by sequencing the same molecule multiple times.
- Uniform Coverage: No bias based on GC content enables sequencing through regions inaccessible to other technologies. Template preparation and sequencing do not rely on amplification, thus there is no PCR bias for more uniform genome coverage.
- Epigenetics: With no PCR amplification step, base modifications are directly detected during sequencing without the need for bisulfite conversion.
Sample requirements
PacBio workflows are sensitive to quality and quantity of input material. It is important to avoid steps in sample extraction and storage that may cause mechanical shearing, fragmentation, or degradation of high molecular weight (HMW) gDNA. Moreover, PacBio is an amplification-free platform, meaning higher input mass is often required for library prep. UMGC performs a full battery of QC on all PacBio submissions. Only samples passing rigorous QC are taken into library preparation. The table below lists guidelines for minimum and recommended sample input:
- Amplicons - 1-10 ng
- Genomic DNA (higher organism) - 5-10 µg
- Genomic DNA (microbial) - 0.5-5 µg
- Full-length (poly-A) mRNA - >300 ng, RIN ≥ 8.0
Questions?
As a PacBio Certified Service Provider with five years of experience with the Sequel Systems, we undergo standardized PacBio certification and speak at national PacBio user group meetings to ensure investigators receive the highest quality data and the longest reads possible. Please contact us at next-gen@umn.edu for comprehensive support on experimental design.

UMN Rates
| Library Creation | Savings Tier | Speed Tier | Priority Tier |
|---|---|---|---|
| Amplicon. No Shearing/Size Selection. | $519.72 | $789.45 | $1,292.68 |
| Turnaround time. | 10 - 15 days | 5 - 10 days | 2 - 4 days |
| HiFi. TMK 3.0 Microbial Multiplexing. | $460.27 | $644.51 | $960.42 |
| Turnaround time. | 12 - 21 days | 7 - 15 days | 3 - 7 days |
| HiFi. TMK 3.0. Includes Library QC. | $849.32 | $1,296.72 | $1,854.68 |
| Turnaround time. | 13 - 21 days | 7 - 14 days | 3 - 7 days |
| IsoSeq.TMK 3.0. Includes Library QC. | $614.31 | $954.31 | $1,272.35 |
| Turnaround time. | 12 - 22 days | 7 - 13 days | 4 - 7 days |
| Sequencing | Savings Tier | Speed Tier | Priority Tier |
|---|---|---|---|
| Revio SMRT Cell 25M. Outsourced Run. | $2,174.01 | $2,704.80 | N/A |
| Turnaround time. | 3 - 10 days | 3 - 6 days | N/A |
| Sequel II SMRT Cell 8M. | $2,041.59 | N/A | N/A |
| Turnaround time. | 3 - 12 days | N/A | N/A |
Notes
Tiers: Tiers: Turnaround time (TAT) is in business days. Availability for the Priority Tier is limited; contact next-gen@umn.edu to see if this is a current option. More information about Pricing Tiers.
External Rates
| Library Creation | Savings Tier | Speed Tier | Priority Tier |
|---|---|---|---|
| Amplicon. No Shearing/Size Selection. | $642.66 | $942.26 | $1,501.19 |
| Turnaround time. | 10 - 15 days | 5 - 10 days | 2 - 4 days |
| HiFi. TMK 3.0 Microbial Multiplexing. | $569.16 | $773.92 | $1,124.81 |
| Turnaround time. | 12 - 21 days | 7 - 15 days | 3 - 7 days |
| HiFi. TMK 3.0. Includes Library QC. | $1,050.25 | $1,547.66 | $2,167.48 |
| Turnaround time. | 13 - 21 days | 7 - 14 days | 3 - 7 days |
| IsoSeq.TMK 3.0. Includes Library QC. | $759.65 | $1,137.42 | $1,490.76 |
| Turnaround time. | 12 - 22 days | 7 - 13 days | 4 - 7 days |
| Sequencing | Savings Tier | Speed Tier | Priority Tier |
|---|---|---|---|
| Revio SMRT Cell 25M. Outsourced Run. | $2,688.30 | $3,277.25 | N/A |
| Turnaround time. | 3 - 10 days | 3 - 6 days | N/A |
| Sequel II SMRT Cell 8M. | $2,524.55 | N/A | N/A |
| Turnaround time. | 3 - 12 days | N/A | N/A |
Notes
Notes
Tiers: Tiers: Turnaround time (TAT) is in business days. Availability for the Priority Tier is limited; contact next-gen@umn.edu to see if this is a current option. More information about Pricing Tiers.
Submission
How to Order
- Please contact next-gen@umn.edu for project specifications.
- After project details are finalized, complete PacBio Sample Submission Form and email to next-gen@umn.edu.
Samples can be dropped off at any of our campus locations
- 1-210 Cancer & Cardiovascular Research Building (Minneapolis campus)
- 20 Snyder Hall (St. Paul campus)
Shipping Instructions
Please give advance notice of your sample submission date and time so staff can be prepared to receive samples. If shipping samples from outside the University of Minnesota, ship via express shipping carrier to the address below.
Please send the tracking information to next-gen@umn.edu.
UMN Genomics Center
ATTN: Elyse Froehling
3510 Hopkins Place N.
Building 4 Suite W402
Oakdale, MN 55128
612-625-7736
Deliverables
Data Release
There are four options for transferring data from the UMGC to clients: 1) delivery to the Minnesota Supercomputing Institute’s (MSI) high-performance file system, 2) download from a secure website, 3) download with Globus, or 4) shipment on an external hard drive. Please indicate your data delivery preference when placing an order for sequencing.
1. MSI storage
Internal clients have their data released to MSI's Shared User Resource Facility Storage (SURFS). Delivered data will be located in the "data_delivery" folder in your group's folder on MSI's primary filesystem (home/GROUP/data_delivery/umgc). MSI does not charge for SURFS storage costs, but files expire and are removed one year after they've been delivered. Files should be copied to other MSI storage locations such as Tier2, Tier3, or your group's "shared" folder before they expire.
2. Web download
Internal clients that opt-out of MSI storage and external clients can download their data from a secure website using either a web browser or a command-line download tool, complete instructions are provided in an email from the UMGC. The client’s data is available for download for 30 days, after which the data will be removed from the data download website and the client takes responsibility for storing the data.
3. Globus
Internal and External clients can use Globus to download their data. This is the recommended method for external clients to download large datasets.
4. Hard drive
External clients may have data shipped on a hard drive purchased by the UMGC and invoiced to the client at a cost of $250 per hard drive.
Data Recovery
The UMGC archives most customer data for a year and some datasets are retained for 5 years or more. If you need a dataset re-delivered email a request to next-gen@umn.edu to initiate data recovery. The UMGC does not provide any guarantee that data can be successfully recovered from the archive.
