As of Tuesday, January 2, 2024, the UMGC is no longer accepting new projects on the NovaSeq 6000, and researchers needing high-output sequencing should use the NovaSeq X Plus or other sequencing options. The NovaSeq 6000 is retiring from our suite of sequencing instruments and returning to the UMN Medical School for critical clinical sequencing. Contact next-gen@umn.edu with questions.
The NovaSeq 6000 is a production-scale sequencer from Illumina that generates unprecedented output in less than two days. The main technical advancements include:
- Patterned flow cells with scalable throughputs to accommodate most projects
- 2-channel chemistry for faster sequencing
- Reduced imaging and data processing time
The patterned flow cells have a defined, organized array of etched wells that each contain a DNA probe for adhering complementary sequences added to DNA libraries. This patterned flow cell replaces the randomly scattered clusters found in imaging-intensive, non-patterned flow cells responsible for slower processing time.
The instrument utilizes Illumina’s 2-channel sequencing by synthesis chemistry, which requires only two images per cycle, instead of the original 4-channel chemistry used by its predecessor, the HiSeq 2500. This simplified nucleotide detection reduces imaging and accelerates data processing times while preserving the same accuracy and quality found in the 4-channel chemistry.
Additional technical details on the NovaSeq instrument can be found on the Illumina website.
Output
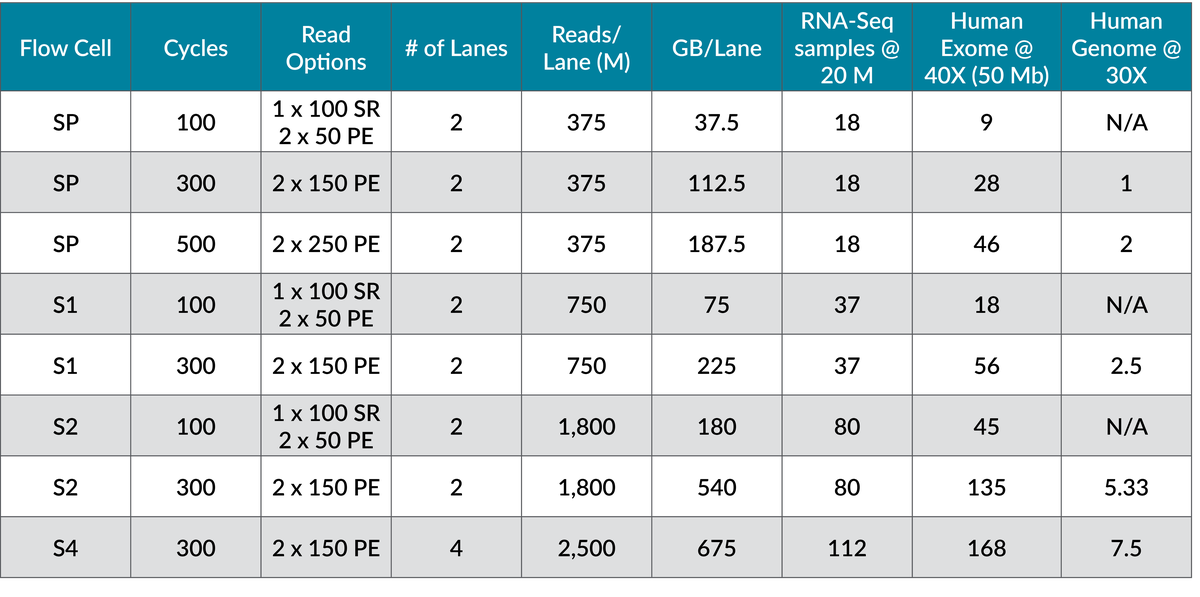
The Genomics Center offers all four NovaSeq flow cell options (SP, S1, S2, and the production-scale S4) as well as the XP protocol, which allows for the loading of individual flow cell lanes with different library types, thereby expanding flow cell flexibility. See a summary of our high-output sequencing options.
Benefits
The NovaSeq produces record output per flow cell combined with reduced on-instrument sequencing durations, thus dramatically improving throughput and speed over previous generations of sequencers. The immense scalability and range of applications for the NovaSeq allows for efficient sequencing of whole human, plant, and animal genomes, whole exomes, and large whole transcriptome projects at the lowest per sample costs compared to other Illumina instruments.
Drawbacks
Careful consideration of sample numbers and flow cell output is crucial for effective use of the platform. The larger capacity NovaSeq may take longer to fill for certain read types, which may delay sequencing turnaround.
Questions
Please contact Aaron Becker or Elyse Cooper at next-gen@umn.edu before submitting a project for the NovaSeq platform.
As of Tuesday, January 2, 2024, the UMGC is no longer accepting new projects on the NovaSeq 6000, and researchers needing high-output sequencing should use the NovaSeq X Plus or other sequencing options. The NovaSeq 6000 is retiring from our suite of sequencing instruments and returning to the UMN Medical School for critical clinical sequencing. Contact next-gen@umn.edu with questions.
Submission
How to order
Researchers can download the appropriate submission form for submitting samples or submitting client-made libraries. Once the form has been completed, please submit the submission by emailing next-gen@umn.edu.
Forms
Illumina Sequencing Request Form (Libraries)
Illumina Sequencing Request Form (Samples)
Samples can be dropped off at one of our campus locations
1-210 Cancer & Cardiovascular Research Building (Minneapolis campus)
20 Snyder Hall (St. Paul campus)
Samples can be shipped to the following address:
Please send the tracking information to next-gen@umn.edu.
UMN Genomics Center
ATTN: NGS Staff
3510 Hopkins Place N.
Building 4 Suite W402
Oakdale, MN 55128
612-625-7736
Deliverables
Data Release
There are four options for transferring data from the UMGC to clients: 1) delivery to the Minnesota Supercomputing Institute’s (MSI) high-performance file system, 2) download from a secure website, 3) download with Globus, or 4) shipment on an external hard drive. Please indicate your data delivery preference when placing an order for sequencing.
1. MSI storage
Internal clients have their data released to MSI's Shared User Resource Facility Storage (SURFS). Delivered data will be located in the "data_delivery" folder in your group's folder on MSI's primary filesystem (home/GROUP/data_delivery/umgc). MSI does not charge for SURFS storage costs, but files expire and are removed one year after they've been delivered. Files should be copied to other MSI storage locations such as Tier2, Tier3, or your group's "shared" folder before they expire.
2. Web download
Internal clients that opt-out of MSI storage and external clients can download their data from a secure website using either a web browser or a command-line download tool, complete instructions are provided in an email from the UMGC. The client’s data is available for download for 30 days, after which the data will be removed from the data download website and the client takes responsibility for storing the data.
3. Globus
Internal and External clients can use Globus to download their data. This is the recommended method for external clients to download large datasets.
4. Hard drive
External clients may have data shipped on a hard drive purchased by the UMGC and invoiced to the client at a cost of $250 per hard drive.
Data Recovery
The UMGC archives most customer data for a year and some datasets are retained for 5 years or more. If you need a dataset re-delivered email a request to next-gen@umn.edu to initiate data recovery. The UMGC does not provide any guarantee that data can be successfully recovered from the archive.
FAQ
Do you accept client-made libraries?
Yes, please consult with UMGC staff before submitting client-made libraries by emailing next-gen@umn.edu.
We recommend that pooling be done at UMGC as PicoGreen concentrations are used for balancing libraries. However, clients may choose to pool themselves.
Client-made libraries will have the following services performed on them before proceeding with sequencing.
- PicoGreen (concentration of library)
- DNA Agilent High Sensitivity chip (size of library)
- KAPA QC (functionality of library)
Where should samples be shipped?
UMN Genomics Center
ATTN: NGS Staff
3510 Hopkins Place N.
Building 4 Suite W402
Oakdale, MN 55128
612-625-7736