Tracking COVID in wastewater: A UMN Genomics Center and Met Council partnership
Wastewater Testing
The UMN Genomics Center (UMGC) and Metropolitan Council scientists have developed an adaptive method for detecting COVID-19 variants in wastewater by identifying the presence of key mutations using ddPCR. By tracking the prevalence of mutations over time, scientists are able to determine probable variants contributing to COVID-19 case surges, including N501Y (Alpha, Beta, & Gamma), L452R (Delta), and K417N (Omicron) variants.
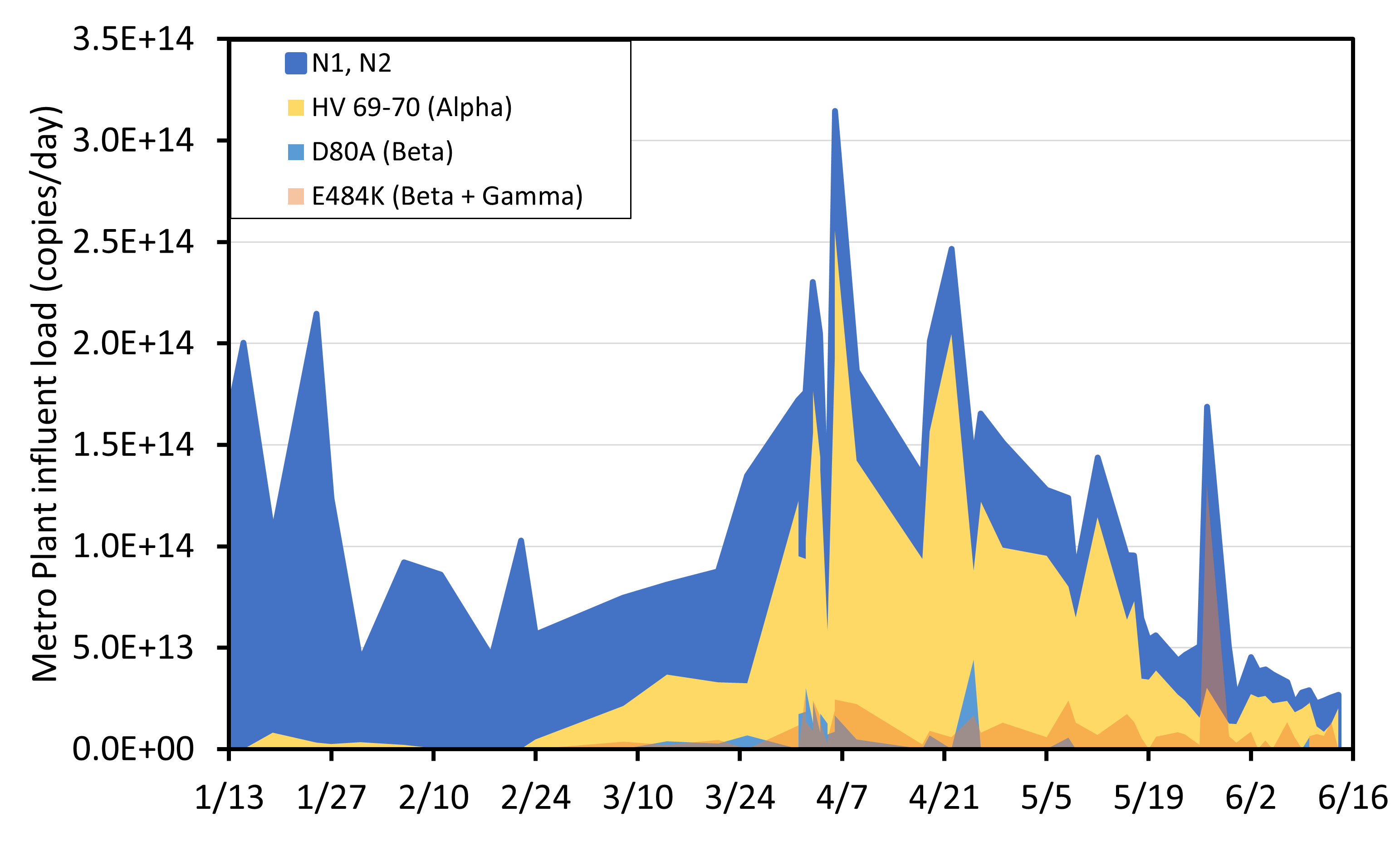
Figure 1: Metro Plant influent load of SARS-COV-2 viral material (N1/N2 gene copies per day) and relative proportion of the mutant allele (variant) at loci associated with variants of concern in wastewater samples. Data was collected in 2021 in partnership with the Metropolitan Council.
COVID Surveillance on Other Fronts
As another way to watch for new strains of SARS-CoV-2, the UMGC has received funding from the CDC to conduct genome sequencing on 6,000 COVID-19 samples to aid national and global viral surveillance efforts. Under the award, UMGC will collaborate with the Minnesota Department of Health to identify which samples to sequence, and then provide the resulting data and analysis from those samples.
In addition to sequencing and wastewater testing, the UMGC has performed hundreds of thousands of COVID-19 tests over the course of the pandemic in collaboration with the UMN Medical School. As employers and schools look to return to in-person working and learning, the center is equipped to provide testing support to these and other organizations that need large-scale, low-cost testing.
Contact the UMGC Director, Dr. Kenny Beckman at kbeckman@umn.edu, with inquiries about COVID services available at the UMGC. For more information on genome sequencing and COVID testing, see the UMGC's SARS-CoV-2 Service page.
